Overview of PyTrial
Table of Contents
As the first chapter of the tutorial, we want to deliver the basic idea of how we design PyTrial in order to help deploy artificial intelligence (AI) algorithms in the real-world drug development process.
Principle: Input & Output Define the Task
We believe the top priority is to ensure that PyTrial is easy-to-use. We are committed to developing a pipeline for each task with a couple lines of codes, for example,
# import the model
from pytrial.tasks.indiv_outcome.tabular import transtab
# initialize the model
model = transtab()
# fit the model
model.fit(train_data, valid_data)
# predict the outcome
model.predict(test_data)
# save the model to disk
model.save_model('./checkpoints')
Without vagueness, this pipeline has an intuitive logic for all machine learning people. We try to keep this interface as simple as possible and its style consistent across all tasks and models. That is to say,
all models should have the same common functions like fit, predict, save_model, load_model, etc. In this way, users can easily switch between different tasks and models without any extra effort to study the interface.
The only thing that remains to care about is then the input and output of the model,
which defines the task.
Code Hierarchy
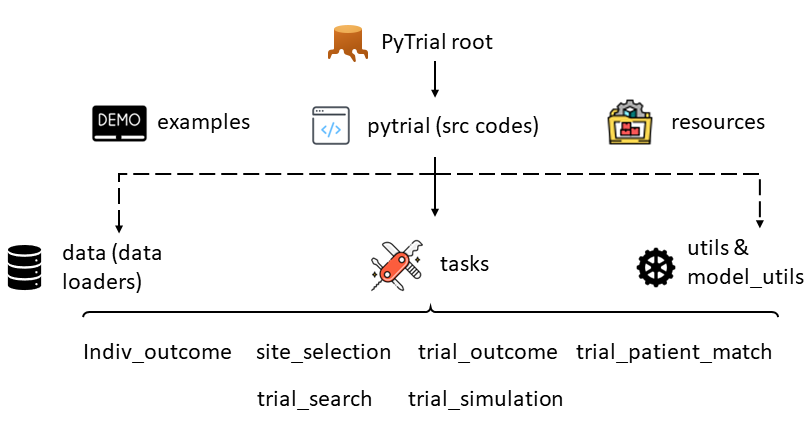
The hierarchy of PyTrial source codes, corresponds to the one in https://github.com/RyanWangZf/PyTrial.
The hierarchy of the PyTrial repository is illustrated by the above figure. On the first layer,
we have the pytrial folder, which contains all the source codes of PyTrial. The other two are the examples that store
the jupyter notebook examples for each model and the resources folder that stores some tools and preprocessed data.
In most scenarios, users only need to use the pytrial folder. The examples folder is for users who want to learn how to use PyTrial. The resources folder is for developers who want to add new models or tasks to PyTrial.
Specifically, the pytrial folder contains the following subfolders:
indiv_outcome: contains the source codes for individualized outcome prediction tasks.
site_selection: contains the source codes for site selection tasks.
trial_outcome: contains the source codes for trial outcome prediction tasks.
trial_patient_match: contains the source codes for trial patient matching tasks.
trial_search: contains the source codes for trial search tasks.
trial_simulation: contains the source codes for trial patient record simulation tasks.
We will play with the models under each task folder, e.g., trial_search.trial2vec. In some cases,
the task has its specific input format, for instance, the patient-trial matching task requires input patient and
trial data in the form of trial_patient_match.data.PatientData and trial_patient_match.data.TrialData, respectively.
Don’t worry, we will go through these tasks one by one with concrete examples!